A College of Liverpool examine has used superior genetic and genomic methods to supply a serious step ahead in understanding and diagnosing infectious intestinal ailments. The analysis is printed within the journal Genome Drugs.
The large-scale examine analyzed greater than 1,000 stool samples from folks with diarrheal sickness to harness two cutting-edge instruments.
Diarrhea, a standard symptom of infectious intestinal illness, impacts an estimated 18 million folks every year within the UK. Nevertheless, regardless of its prevalence, conventional lab checks typically fail to acknowledge its trigger, particularly when infections are attributable to unidentifiable or rising pathogens.
The examine used metagenomic (DNA-based) and metatranscriptomic (gene or RNA-based) sequencing. Not like conventional strategies, these methods don’t depend on rising organisms in a lab. As a substitute, they detect and analyze the genetic materials instantly from affected person samples.
Co-lead writer, Dr. Edward Cunningham-Oakes, Institute of An infection, Veterinary and Ecological Sciences, College of Liverpool mentioned, “That is the UK’s largest examine to check conventional diagnostics with these next-generation instruments. We not solely discovered infections missed by normal checks, however we might see what the bugs had been doing contained in the intestine—one thing normal diagnostics simply cannot present.”
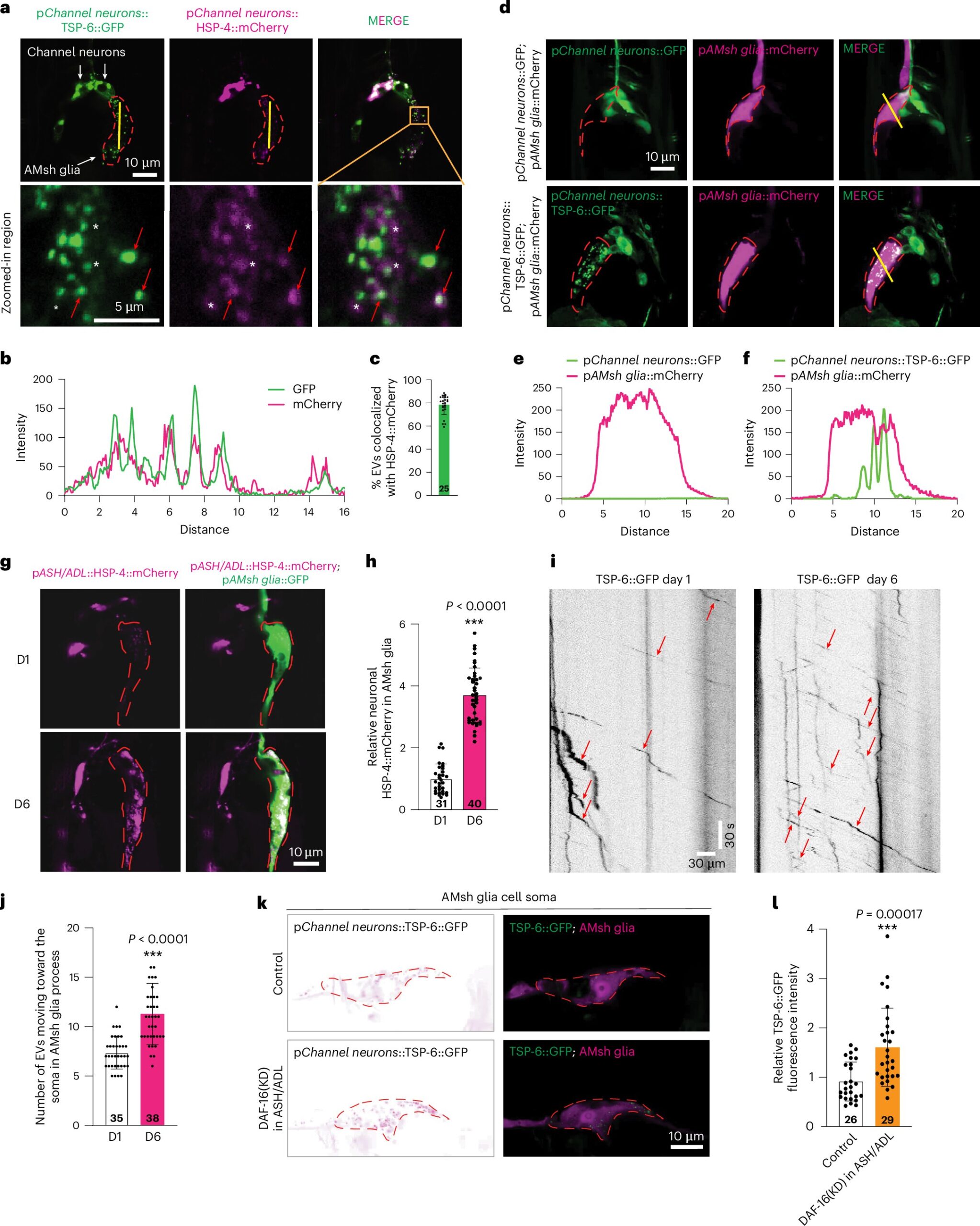
The work captures, for the primary time, a complete snapshot of the Salmonella gene expression instantly from a human stool pattern. The transcriptomic information gives new insights into how the micro organism survive and adapt after leaving the human intestine.
As Salmonella stays a precedence diarrheal pathogen within the UK, the data might be invaluable for serving to scientists to focus on this harmful pathogen.
The examine’s key findings spotlight the facility of RNA checks in detecting hidden infections—together with elusive parasites and RNA viruses—whereas additionally figuring out which genes are lively throughout an infection. Remarkably, RNA remained steady in stool samples even with out preservatives, suggesting it’s extra sturdy than beforehand thought.
Notably, the ratio of RNA to DNA helped differentiate true infections from innocent intestine microbes. By combining each DNA and RNA information, researchers gained the clearest and most correct image of the an infection course of.
Co-lead Professor Alistair Darby, Co-Director of the College’s Heart for Genomic Analysis, mentioned, “This examine reveals how genetic instruments can revolutionize how we establish and perceive intestinal infections. By understanding not simply what’s there, however what it is doing, we are able to enhance public well being responses, significantly round food-borne outbreaks.”
The findings might considerably impression how diarrheal ailments are identified, managed, and studied within the UK and past—particularly in gentle of the rising want for speedy, correct diagnostics that do not depend on outdated culturing strategies.
The analysis additionally highlights the strategic function of Liverpool’s Heart for Genomic Analysis (CGR) and the brand new Microbiome and Infectious Illness (MaID) initiative, which is a part of the Liverpool Metropolis Area’s Life Sciences Innovation Zone.
Professor Alistair Darby continued, “That is about greater than diagnosing infections—it is about constructing a platform for innovation in well being care. Our earlier work has proven that well being care professionals are open to this. By making our information open-access, we hope to assist different researchers, NHS labs, and public well being businesses construct on our work.”
Dr. Edward Cunningham-Oakes added, “Our outcomes present that RNA, as soon as thought too fragile to make use of in stool testing, can truly give us highly effective insights into how infections work. That opens up new prospects for diagnosing and treating these sicknesses extra successfully.”
Extra info:
Edward Cunningham-Oakes et al, Enhancing infectious intestinal illness prognosis via metagenomic and metatranscriptomic sequencing of 1000 human diarrhoeal samples, Genome Drugs (2025). DOI: 10.1186/s13073-025-01478-w
Quotation:
Superior genomics examine improves detection of hard-to-find diarrheal infections (2025, Could 20)
retrieved 20 Could 2025
from https://medicalxpress.com/information/2025-05-advanced-genomics-hard-diarrheal-infections.html
This doc is topic to copyright. Other than any honest dealing for the aim of personal examine or analysis, no
half could also be reproduced with out the written permission. The content material is offered for info functions solely.